Note
This is the documentation for the current state of the development branch of Qiskit Experiments. The documentation or APIs here can change prior to being released.
Getting Started¶
Installation¶
Qiskit Experiments is built on top of Qiskit, so we recommend that you first install Qiskit following its installation guide. Qiskit Experiments supports the same platforms as Qiskit itself and Python versions 3.10 through 3.14.
Qiskit Experiments releases can be installed via the Python package manager pip
in your shell environment:
python -m pip install qiskit-experiments
There are a number of optional packages that enable some experiments and features. If you would like to install these optional dependencies, run:
python -m pip install "qiskit-experiments[extras]"
If you want to run the most up-to-date version instead (may not be stable), you can install the latest main branch:
python -m pip install git+https://github.com/Qiskit-Community/qiskit-experiments.git
If you want to develop the package, you can install Qiskit Experiments from source by cloning the repository:
git clone https://github.com/Qiskit-Community/qiskit-experiments.git
python -m pip install -e "qiskit-experiments[extras]"
The -e option will keep your installed package up to date as you make or pull new
changes. See also the contributing document
in the source code repository for more information on developing the package.
Upgrading Qiskit Experiments¶
Qiskit Experiments version numbers are in the form 0.X.Y, where X is the minor version and
Y is the patch version. There are two kinds of releases: minor releases, which increment the
minor version, and patch releases, which increment the patch version. New features and API
changes can only be introduced in a minor release. Patch releases contain only bug fixes and changes that do
not affect how you use the package, such as performance optimization and documentation updates.
Therefore, when you encounter a bug or unexpected behavior, it is recommended that you first check if there’s a
patch release you can upgrade to under the same minor version to avoid any breaking changes. When
running pip, you can specify the exact version to install:
python -m pip install qiskit-experiments==0.X.Y
Before a nontrivial breaking API change is introduced in a minor release, the old feature will undergo a deprecation process lasting two releases for a core framework change and one release otherwise. During this process, deprecation warnings will be issued if you use the old feature that will instruct you on how to transition to the replacement feature, if applicable. The release notes contain full details on which features are deprecated or removed in each release.
Running your first experiment¶
Let’s run a T1 Experiment, which estimates the characteristic relaxation time
of a qubit from the excited state to the ground state, also known as \(T_1\), by
measuring the excited state population after varying delays. First, we have to import
the experiment from the Qiskit Experiments library:
from qiskit_experiments.library import T1
Experiments must be run on a backend. We’re going to use a simulator,
FakePerth, for this example, but you can use any
backend, real or simulated, that you can access through Qiskit.
Note
This tutorial requires the qiskit-aer and qiskit-ibm-runtime
packages to run simulations. You can install them with python -m pip
install qiskit-aer qiskit-ibm-runtime.
from qiskit_ibm_runtime.fake_provider import FakePerth
from qiskit_aer import AerSimulator
backend = AerSimulator.from_backend(FakePerth())
All experiments require a physical_qubits parameter as input that specifies which
physical qubit or qubits the circuits will be executed on. The qubits must be given as a
Python sequence (usually a tuple or a list).
In addition, the \(T_1\) experiment has
a second required parameter, delays, which is a list of times in seconds at which to
measure the excited state population. In this example, we’ll run the \(T_1\)
experiment on qubit 0, and use the t1 backend property of this qubit to give us a
good estimate for the sweep range of the delays.
import numpy as np
qubit0_t1 = FakePerth().qubit_properties(0).t1
delays = np.arange(1e-6, 3 * qubit0_t1, 3e-5)
exp = T1(physical_qubits=(0,), delays=delays)
The circuits encapsulated by the experiment can be accessed using the experiment’s
circuits() method, which returns a list of circuits that can be
run on a backend. Let’s print the range of delay times we’re sweeping over and draw the
first and last circuits for our \(T_1\) experiment:
print(delays)
exp.circuits()[0].draw(output="mpl", style="iqp")
[1.00e-06 3.10e-05 6.10e-05 9.10e-05 1.21e-04 1.51e-04]
exp.circuits()[-1].draw(output="mpl", style="iqp")
As expected, the delay block spans the full range of time values that we specified.
The ExperimentData class¶
After instantiating the experiment, we run the experiment by calling
run() with our backend of choice. This transpiles our experiment
circuits then packages them into jobs that are run on the backend.
Note
See the how-tos for customizing job splitting when running an experiment.
This statement returns the ExperimentData class containing the results of the
experiment, so it’s crucial that we assign the output to a data variable. We could have
also provided the backend at the instantiation of the experiment, but specifying the
backend at run time allows us to run the same exact experiment on different backends
should we choose to do so.
exp_data = exp.run(backend=backend).block_for_results()
The block_for_results() method is optional and is used to block
execution of subsequent code until the experiment has fully completed execution and
analysis. If
exp_data = exp.run(backend=backend)
is executed instead, the statement will finish running as soon as the jobs are
submitted, but the analysis callback won’t populate exp_data with results until the
entire process has finished. In this case, there are two useful methods in the
ExperimentData, job_status() and
analysis_status(), that return the current status of the job and
analysis, respectively:
print(exp_data.job_status())
print(exp_data.analysis_status())
JobStatus.DONE
AnalysisStatus.DONE
Figures¶
Once the analysis is complete, figures are retrieved using the
figure() method. See the visualization module tutorial on how to customize figures for an experiment. For our
\(T_1\) experiment, we have a single figure showing the raw data and fit to the
exponential decay model of the \(T_1\) experiment:
display(exp_data.figure(0))
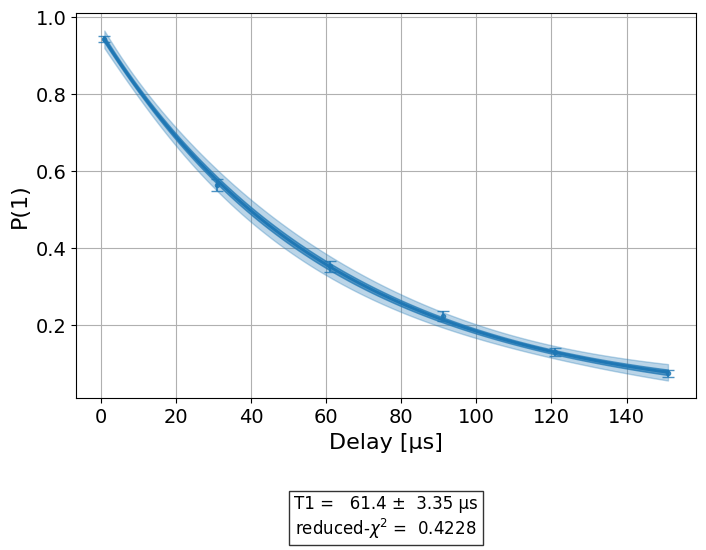
Analysis Results¶
The analysis results resulting from the fit are accessed with
analysis_results(). If the dataframe=True parameter is passed, analysis
results will be retrieved in the pandas DataFrame format:
exp_data.analysis_results(dataframe=True)
| name | experiment | components | value | quality | backend | run_time | chisq | unit | |
|---|---|---|---|---|---|---|---|---|---|
| dc3be86b | T1 | T1 | [Q0] | (5.18+/-0.27)e-05 | good | aer_simulator_from(fake_perth) | None | 0.299338 | s |
Each analysis
result value is a UFloat object from the uncertainties package. The nominal
value and standard deviation of each value can be accessed as follows:
t1 = exp_data.analysis_results("T1", dataframe=True).iloc[0]
print(t1.value.nominal_value)
print(t1.value.std_dev)
5.181075051051895e-05
2.6905558395986928e-06
For further documentation on how to work with UFloats, consult the uncertainties
User Guide.
Artifacts¶
The curve fit data itself is contained in artifacts(), which are accessed
in an analogous manner. Artifacts for a standard experiment include both the curve fit data
stored in artifacts("curve_data") and information on the fit stored in artifacts("fit_summary").
Use the data attribute to access artifact data:
print(exp_data.artifacts("fit_summary").data)
CurveFitResult:
- fitting method: least_squares
- number of sub-models: 1
* F_exp_decay(x) = amp * exp(-x/tau) + base
- success: True
- number of function evals: 25
- degree of freedom: 3
- chi-square: 0.8980128454418626
- reduced chi-square: 0.2993376151472875
- Akaike info crit.: -5.395982253056042
- Bayesian info crit.: -6.020703845371877
- init params:
* amp = 0.8653658536585366
* tau = 6.583423621282744e-05
* base = 0.09707317073170732
- fit params:
* amp = 0.929436371534897 ± 0.014933586311156873
* tau = 5.181075051051895e-05 ± 2.6905558395986928e-06
* base = 0.05039549777033997 ± 0.014433837329511076
- correlations:
* (tau, base) = -0.8916622874144673
* (amp, base) = -0.8687907821757553
* (amp, tau) = 0.7025198883027572
Note
See the artifacts how-to for more information on using artifacts.
Circuit data and metadata¶
Raw circuit output data and its associated metadata can be accessed with the
data() property. Data is indexed by the circuit it corresponds
to. Depending on the measurement level set in the experiment, the raw data will either
be in the key counts (level 2) or memory (level 1 IQ data).
Note
See the data processor tutorial for more information on level 1 and level 2 data.
Circuit metadata contains information set by the experiment on a circuit-by-circuit
basis; xval is used by the analysis to extract the x value for each circuit when
fitting the data.
print(exp_data.data(0))
{'job_id': '36e54d8a-04ef-4b78-b4c5-2a7f73fbea9a', 'meas_level': 2, 'meas_return': 'single', 'counts': {'1': 986, '0': 38}, 'memory': ['1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1', '1', '1', '1', '1', '0', '1', '1', '1', '1'], 'shots': 1024, 'metadata': {'xval': 9.99111111111111e-07}}
Experiments also have global associated metadata accessed by the
metadata() property.
print(exp_data.metadata)
{'physical_qubits': [0], 'device_components': [<Qubit(Q0)>], 'meas_level': <MeasLevel.CLASSIFIED: 2>, '_source': {'class': 'qiskit_experiments.framework.experiment_data.ExperimentData', 'metadata_version': 1, 'qiskit_version': {'qiskit': '2.3.0', 'qiskit-experiments': '0.14.0'}}}
Job information¶
The actual backend jobs that were executed for the experiment can be accessed with the
jobs() method.
Note
See the how-tos for rerunning the analysis for an existing experiment that finished execution.
Setting options for your experiment¶
It’s often insufficient to run an experiment with only its default options. There are four types of options one can set for an experiment:
Run options¶
These options are passed to the experiment’s run() method and
then to the run() method of your specified backend. Any run option that your backend
supports can be set:
from qiskit_experiments.framework import MeasLevel
exp.set_run_options(shots=1000,
meas_level=MeasLevel.CLASSIFIED)
print(f"Shots set to {exp.run_options.get('shots')}, "
"measurement level set to {exp.run_options.get('meas_level')}")
Shots set to 1000, measurement level set to {exp.run_options.get('meas_level')}
Consult the documentation of the run method of your
specific backend type for valid options.
For example, see qiskit_ibm_runtime.IBMBackend.run() for IBM backends.
Transpile options¶
These options are passed to the Qiskit transpiler to transpile the experiment circuits
before execution:
exp.set_transpile_options(scheduling_method='asap',
optimization_level=3,
basis_gates=["x", "sx", "rz"])
print(f"Transpile options are {exp.transpile_options}")
Transpile options are Options(optimization_level=3, scheduling_method='asap', basis_gates=['x', 'sx', 'rz'])
Consult the documentation of qiskit.compiler.transpile() for valid options.
Experiment options¶
These options are unique to each experiment class. Many experiment options can be set
upon experiment instantiation, but can also be explicitly set via
set_experiment_options():
exp = T1(physical_qubits=(0,), delays=delays)
new_delays=np.arange(1e-6, 600e-6, 50e-6)
exp.set_experiment_options(delays=new_delays)
print(f"Experiment options are {exp.experiment_options}")
Experiment options are Options(max_circuits=None, delays=array([1.00e-06, 5.10e-05, 1.01e-04, 1.51e-04, 2.01e-04, 2.51e-04,
3.01e-04, 3.51e-04, 4.01e-04, 4.51e-04, 5.01e-04, 5.51e-04]))
Consult the API documentation for the options of each experiment class.
Analysis options¶
These options are unique to each analysis class. Unlike the other options, analysis
options are not directly set via the experiment object but use instead a method of the
associated analysis:
from qiskit_experiments.library import StandardRB
exp = StandardRB(physical_qubits=(0,),
lengths=list(range(1, 300, 30)),
seed=123,
backend=backend)
exp.analysis.set_options(gate_error_ratio=None)
Consult the API documentation for the options of each experiment’s analysis class.
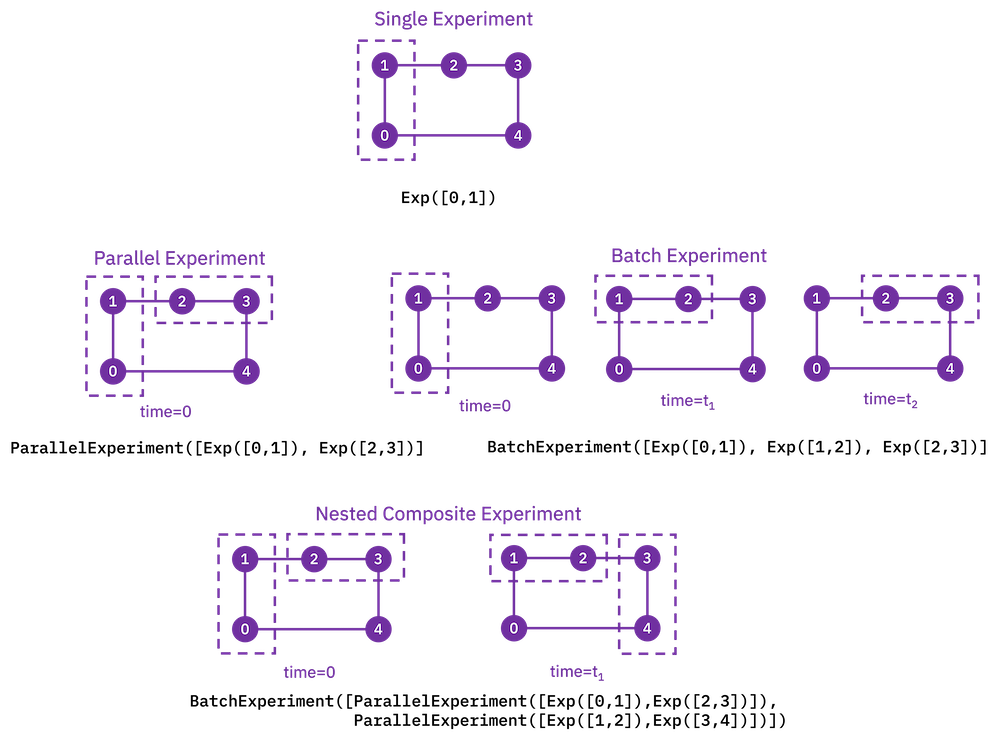
Running experiments on multiple qubits¶
To run experiments across many qubits of the same device, we use composite
experiments. A CompositeExperiment is a parent object that contains one or more child
experiments, which may themselves be composite. There are two core types of composite
experiments:
Parallel experiments run across qubits simultaneously as set by the user. The circuits of child experiments are combined into new circuits that map circuit gates onto qubits in parallel. Therefore, the circuits in child experiments cannot overlap in the
physical_qubitsparameter. The marginalization of measurement data for analysis of each child experiment is handled automatically.Batch experiments run consecutively in time. These child circuits can overlap in qubits used.
Using parallel experiments, we can measure the \(T_1\) of one qubit while doing a
standard Randomized Benchmarking StandardRB experiment on other qubits
simultaneously on the same device:
from qiskit_experiments.framework import ParallelExperiment
child_exp1 = T1(physical_qubits=(2,), delays=delays)
child_exp2 = StandardRB(physical_qubits=(3,1), lengths=np.arange(1,100,10), num_samples=2)
parallel_exp = ParallelExperiment([child_exp1, child_exp2])
Note that when the transpile and run options are set for a composite experiment, the
child experiments’s options are also set to the same options recursively. Let’s examine
how the parallel experiment is constructed by visualizing child and parent circuits. The
child experiments can be accessed via the
component_experiment() method, which indexes from zero:
parallel_exp.component_experiment(0).circuits()[0].draw(output="mpl", style="iqp")
parallel_exp.component_experiment(1).circuits()[0].draw(output="mpl", style="iqp")
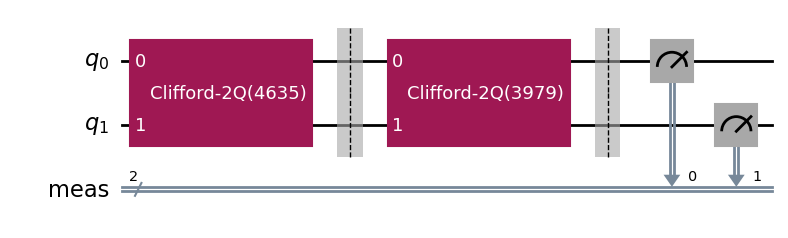
Similarly, the child analyses can be accessed via CompositeAnalysis.component_analysis() or via
the analysis of the child experiment class:
parallel_exp.component_experiment(0).analysis.set_options(plot = True)
# This should print out what we set because it's the same option
print(parallel_exp.analysis.component_analysis(0).options.get("plot"))
True
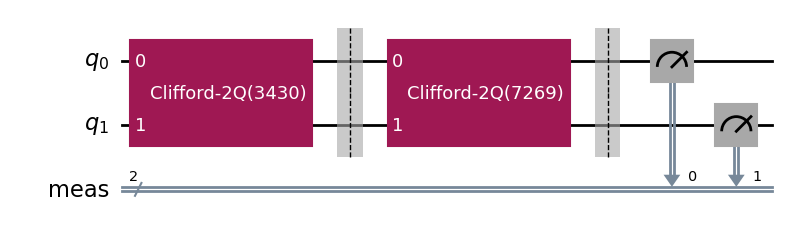
The circuits of all experiments assume they’re acting on virtual qubits starting from index 0. In the case of a parallel experiment, the child experiment circuits are composed together and then reassigned virtual qubit indices:
parallel_exp.circuits()[0].draw(output="mpl", style="iqp")
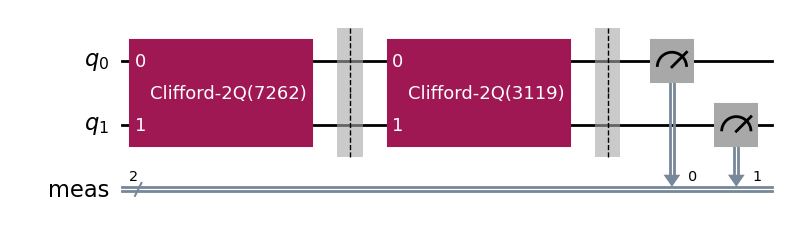
During experiment transpilation, a mapping is performed to place these circuits on the
physical layout. We can see its effects by looking at the transpiled
circuit, which is accessed via the internal method _transpiled_circuits(). After
transpilation, the T1 experiment is correctly placed on physical qubit 2
and the StandardRB experiment’s gates are on physical qubits 3 and 1.
parallel_exp._transpiled_circuits()[0].draw(output="mpl", style="iqp")
ParallelExperiment and BatchExperiment classes can also be nested
arbitrarily to make complex composite experiments.
Viewing child experiment data¶
The experiment data returned from a composite experiment contains analysis results for each child experiment in the parent experiment.
Note
By default, all analysis results will be stored in the parent data object,
and you need to explicitly set flatten_results=False to generate child
data objects in the legacy format.
parallel_exp = ParallelExperiment(
[T1(physical_qubits=(i,), delays=delays) for i in range(2)]
)
parallel_data = parallel_exp.run(backend, seed_simulator=101).block_for_results()
parallel_data.analysis_results(dataframe=True)
| name | experiment | components | value | quality | backend | run_time | chisq | unit | |
|---|---|---|---|---|---|---|---|---|---|
| b4350f40 | T1 | T1 | [Q0] | (6.14+/-0.35)e-05 | good | aer_simulator_from(fake_perth) | None | 0.846896 | s |
| 22c986d7 | T1 | T1 | [Q1] | 0.000129+/-0.000019 | good | aer_simulator_from(fake_perth) | None | 0.477908 | s |
Broadcasting analysis options to child experiments¶
Use the broadcast parameter to set analysis options to each of the child experiments.
parallel_exp.analysis.set_options(plot=False, broadcast=True)
If the child experiment inherits from CompositeExperiment (such as ParallelExperiment
and BatchExperiment classes), this process will continue to work recursively.
In this instance, the analysis will not generate a figure for the child experiment after the analysis.