Quantum State Tomography¶
Quantum tomography is an experimental procedure to reconstruct a description of part of a quantum system from the measurement outcomes of a specific set of experiments. In particular, quantum state tomography reconstructs the density matrix of a quantum state by preparing the state many times and measuring them in a tomographically complete basis of measurement operators.
Note
This tutorial requires the qiskit-aer and qiskit-ibm-runtime
packages to run simulations. You can install them with python -m pip
install qiskit-aer qiskit-ibm-runtime.
We first initialize a simulator to run the experiments on.
from qiskit_aer import AerSimulator
from qiskit_ibm_runtime.fake_provider import FakePerth
backend = AerSimulator.from_backend(FakePerth())
To run a state tomography experiment, we initialize the experiment with a circuit to
prepare the state to be measured. We can also pass in an
Operator or a Statevector
to describe the preparation circuit.
import qiskit
from qiskit_experiments.framework import ParallelExperiment
from qiskit_experiments.library import StateTomography
# GHZ State preparation circuit
nq = 2
qc_ghz = qiskit.QuantumCircuit(nq)
qc_ghz.h(0)
qc_ghz.s(0)
for i in range(1, nq):
qc_ghz.cx(0, i)
# QST Experiment
qstexp1 = StateTomography(qc_ghz)
qstdata1 = qstexp1.run(backend, seed_simulation=100).block_for_results()
# Print results
display(qstdata1.analysis_results(dataframe=True))
| name | experiment | components | value | quality | backend | run_time | trace | eigvals | raw_eigvals | rescaled_psd | fitter_metadata | conditional_probability | positive | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6c6a05c4 | state | StateTomography | [Q0, Q1] | DensityMatrix([[ 0.45914714+0.j , 0.01... | None | aer_simulator_from(fake_perth) | None | 1.0 | [0.919198156377239, 0.04698324334911811, 0.028... | [0.919198156377239, 0.04698324334911811, 0.028... | False | {'fitter': 'linear_inversion', 'fitter_time': ... | 1.0 | True |
| 469d0d48 | state_fidelity | StateTomography | [Q0, Q1] | 0.918457 | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| 6204da39 | positive | StateTomography | [Q0, Q1] | True | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
Tomography Results¶
The main result for tomography is the fitted state, which is stored as a
DensityMatrix object:
state_result = qstdata1.analysis_results("state", dataframe=True).iloc[0]
print(state_result.value)
DensityMatrix([[ 0.45914714+0.j , 0.01025391-0.0086263j ,
0.00927734+0.00276693j, -0.01318359-0.44921875j],
[ 0.01025391+0.0086263j , 0.03011068+0.j ,
-0.01318359+0.j , -0.01123047-0.00406901j],
[ 0.00927734-0.00276693j, -0.01318359+0.j ,
0.03141276+0.j , -0.00830078-0.01155599j],
[-0.01318359+0.44921875j, -0.01123047+0.00406901j,
-0.00830078+0.01155599j, 0.47932943+0.j ]],
dims=(2, 2))
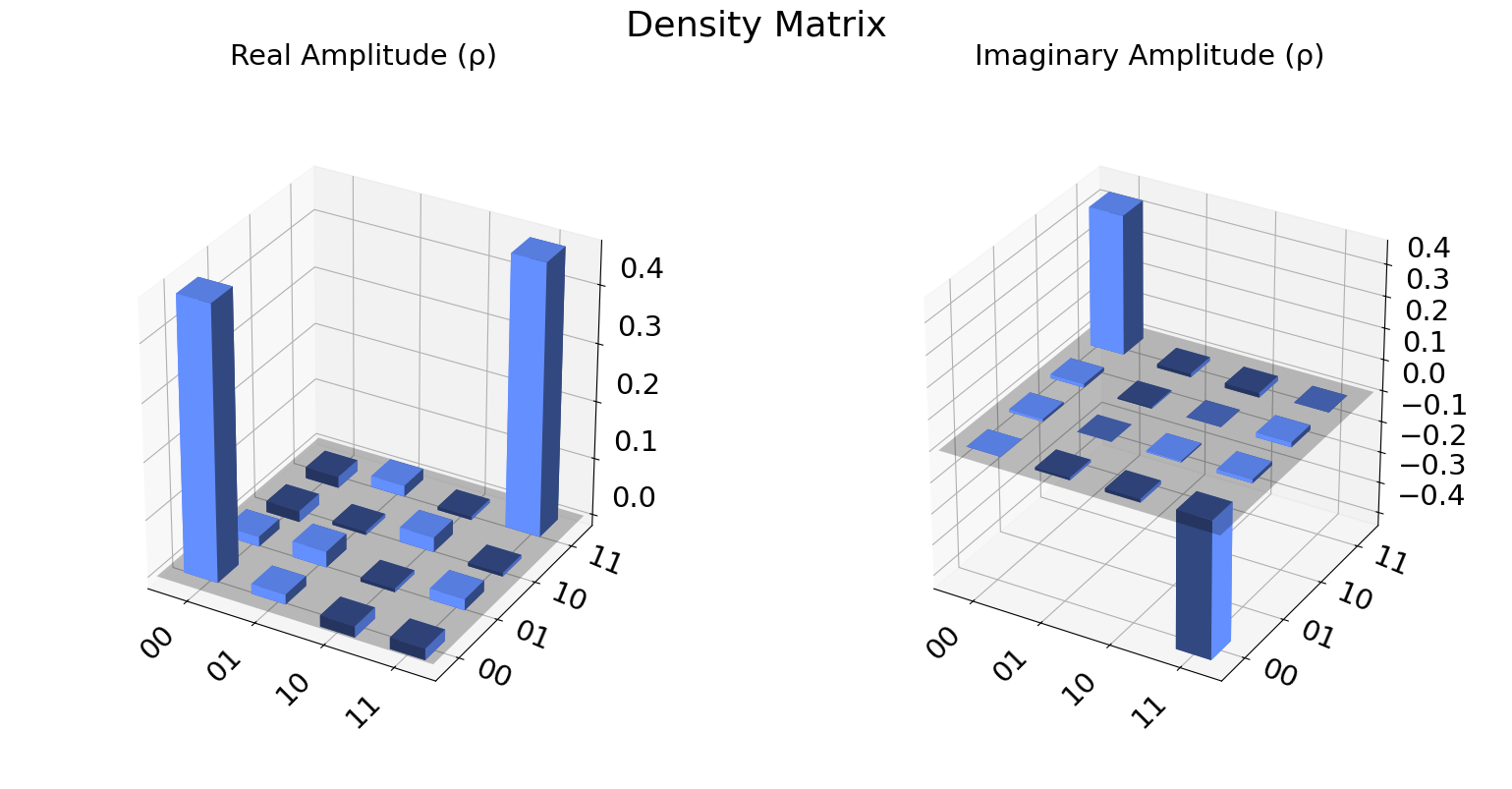
We can also visualize the density matrix:
from qiskit.visualization import plot_state_city
state = qstdata1.analysis_results("state", dataframe=True).iloc[0].value
plot_state_city(state, title='Density Matrix')
The state fidelity of the fitted state with the ideal state prepared by
the input circuit is stored in the "state_fidelity" result field.
Note that if the input circuit contained any measurements the ideal
state cannot be automatically generated and this field will be set to
None.
fid_result = qstdata1.analysis_results("state_fidelity", dataframe=True).iloc[0]
print("State Fidelity = {:.5f}".format(fid_result.value))
State Fidelity = 0.91846
Additional state metadata¶
Additional data is stored in the tomography under additional fields. This includes
eigvals: the eigenvalues of the fitted statetrace: the trace of the fitted statepositive: Whether the eigenvalues are all non-negative
If trace rescaling was performed this dictionary will also contain a raw_trace field
containing the trace before rescaling. Futhermore, if the state was rescaled to be
positive or trace 1 an additional field raw_eigvals will contain the state
eigenvalues before rescaling was performed.
for col in ["eigvals", "trace", "positive"]:
print(f"{col}: {state_result[col]}")
eigvals: [0.91919816 0.04698324 0.02892367 0.00489493]
trace: 1.000000000000001
positive: True
To see the effect of rescaling, we can perform a “bad” fit with very low counts:
# QST Experiment
bad_data = qstexp1.run(backend, shots=10, seed_simulation=100).block_for_results()
bad_state_result = bad_data.analysis_results("state", dataframe=True).iloc[0]
# Print result
for key, val in bad_state_result.items():
print(f"{key}: {val}")
name: state
experiment: StateTomography
components: [<Qubit(Q0)>, <Qubit(Q1)>]
value: DensityMatrix([[ 0.43608661+0.00000000e+00j, 0.09545365+1.29166135e-02j,
-0.03688001+7.32474804e-02j, 0.00646826-3.49857117e-01j],
[ 0.09545365-1.29166135e-02j, 0.03542661+0.00000000e+00j,
-0.00897047+9.70260775e-03j, 0.03263655-1.00678478e-01j],
[-0.03688001-7.32474804e-02j, -0.00897047-9.70260775e-03j,
0.02861705+0.00000000e+00j, -0.07299639+4.16493120e-02j],
[ 0.00646826+3.49857117e-01j, 0.03263655+1.00678478e-01j,
-0.07299639-4.16493120e-02j, 0.49986973-6.93889390e-18j]],
dims=(2, 2))
quality: None
backend: aer_simulator_from(fake_perth)
run_time: None
trace: 1.0000000000000009
eigvals: [0.86026194 0.12496074 0.01477732 0. ]
raw_eigvals: [ 0.89891231 0.16361111 0.05342768 -0.1159511 ]
rescaled_psd: True
fitter_metadata: {'fitter': 'linear_inversion', 'fitter_time': 0.0039331912994384766}
conditional_probability: 1.0
positive: True
Tomography Fitters¶
The default fitters is linear_inversion, which reconstructs the
state using dual basis of the tomography basis. This will typically
result in a non-positive reconstructed state. This state is rescaled to
be positive-semidefinite (PSD) by computing its eigen-decomposition and
rescaling its eigenvalues using the approach from Ref. [1].
There are several other fitters are included (See API documentation for
details). For example, if cvxpy is installed we can use the
cvxpy_gaussian_lstsq() fitter, which allows constraining the fit to be
PSD without requiring rescaling.
try:
import cvxpy
# Set analysis option for cvxpy fitter
qstexp1.analysis.set_options(fitter='cvxpy_gaussian_lstsq')
# Re-run experiment
qstdata2 = qstexp1.run(backend, seed_simulation=100).block_for_results()
state_result2 = qstdata2.analysis_results("state", dataframe=True).iloc[0]
for key, val in state_result2.items():
print(f"{key}: {val}")
except ModuleNotFoundError:
print("CVXPY is not installed")
name: state
experiment: StateTomography
components: [<Qubit(Q0)>, <Qubit(Q1)>]
value: DensityMatrix([[ 4.77385497e-01+0.j , 9.13079993e-03-0.01134232j,
8.33375234e-03+0.0031693j , -1.51344600e-02-0.44016457j],
[ 9.13079993e-03+0.01134232j, 3.51646408e-02+0.j ,
-1.80746266e-02+0.00279125j, -3.66068786e-03-0.00364678j],
[ 8.33375234e-03-0.0031693j , -1.80746266e-02-0.00279125j,
2.21091687e-02+0.j , 3.65148367e-04+0.00783394j],
[-1.51344600e-02+0.44016457j, -3.66068786e-03+0.00364678j,
3.65148367e-04-0.00783394j, 4.65340694e-01+0.j ]],
dims=(2, 2))
quality: None
backend: aer_simulator_from(fake_perth)
run_time: None
trace: 1.000000002034044
eigvals: [0.91196631 0.05425153 0.0300069 0.00377526]
raw_eigvals: [0.91196631 0.05425153 0.0300069 0.00377526]
rescaled_psd: False
fitter_metadata: {'fitter': 'cvxpy_gaussian_lstsq', 'cvxpy_solver': 'SCS', 'cvxpy_status': ['optimal'], 'psd_constraint': True, 'trace_preserving': True, 'fitter_time': 0.03258657455444336}
conditional_probability: 1.0
positive: True
Parallel Tomography Experiment¶
We can also use the ParallelExperiment class to
run subsystem tomography on multiple qubits in parallel.
For example if we want to perform 1-qubit QST on several qubits at once:
from math import pi
num_qubits = 5
gates = [qiskit.circuit.library.RXGate(i * pi / (num_qubits - 1))
for i in range(num_qubits)]
subexps = [
StateTomography(gate, physical_qubits=(i,))
for i, gate in enumerate(gates)
]
parexp = ParallelExperiment(subexps)
pardata = parexp.run(backend, seed_simulation=100).block_for_results()
display(pardata.analysis_results(dataframe=True))
| name | experiment | components | value | quality | backend | run_time | trace | eigvals | raw_eigvals | rescaled_psd | fitter_metadata | conditional_probability | positive | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| caea0fe6 | state | StateTomography | [Q0] | DensityMatrix([[ 0.96972656+0.j , -0.00... | None | aer_simulator_from(fake_perth) | None | 1.0 | [0.9697681813033308, 0.03023181869667016] | [0.9697681813033308, 0.03023181869667016] | False | {'fitter': 'linear_inversion', 'fitter_time': ... | 1.0 | True |
| f849e946 | state_fidelity | StateTomography | [Q0] | 0.969727 | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| 4e995942 | positive | StateTomography | [Q0] | True | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| 8c824478 | state | StateTomography | [Q1] | DensityMatrix([[0.86262789+0.j , 0.0048... | None | aer_simulator_from(fake_perth) | None | 1.0 | [1.0000000000000009, 0.0] | [1.0035938802891162, -0.0035938802891152255] | True | {'fitter': 'linear_inversion', 'fitter_time': ... | 1.0 | True |
| 4bcfb943 | state_fidelity | StateTomography | [Q1] | 0.999807 | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| 60e90429 | positive | StateTomography | [Q1] | True | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| ae0904dd | state | StateTomography | [Q2] | DensityMatrix([[ 0.51171875+0.j , -0.01... | None | aer_simulator_from(fake_perth) | None | 1.0 | [0.9720232800060566, 0.02797671999394402] | [0.9720232800060566, 0.02797671999394402] | False | {'fitter': 'linear_inversion', 'fitter_time': ... | 1.0 | True |
| 9b20b01c | state_fidelity | StateTomography | [Q2] | 0.97168 | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| f12ab20d | positive | StateTomography | [Q2] | True | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| a1fc0e73 | state | StateTomography | [Q3] | DensityMatrix([[ 0.14453125+0.j , -0.006... | None | aer_simulator_from(fake_perth) | None | 1.0 | [0.9891408449253822, 0.010859155074618987] | [0.9891408449253822, 0.010859155074618987] | False | {'fitter': 'linear_inversion', 'fitter_time': ... | 1.0 | True |
| bac92e1e | state_fidelity | StateTomography | [Q3] | 0.988898 | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| 0a945e02 | positive | StateTomography | [Q3] | True | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| c54dc0d2 | state | StateTomography | [Q4] | DensityMatrix([[0.03125 +0.j , 0.0253... | None | aer_simulator_from(fake_perth) | None | 1.0 | [0.9695387244621378, 0.030461275537863288] | [0.9695387244621378, 0.030461275537863288] | False | {'fitter': 'linear_inversion', 'fitter_time': ... | 1.0 | True |
| 420aa95b | state_fidelity | StateTomography | [Q4] | 0.96875 | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| 959f5349 | positive | StateTomography | [Q4] | True | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
View experiment analysis results for one component:
results = pardata.analysis_results(dataframe=True)
display(results[results.components.apply(lambda x: x == ["Q0"])])
| name | experiment | components | value | quality | backend | run_time | trace | eigvals | raw_eigvals | rescaled_psd | fitter_metadata | conditional_probability | positive | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| caea0fe6 | state | StateTomography | [Q0] | DensityMatrix([[ 0.96972656+0.j , -0.00... | None | aer_simulator_from(fake_perth) | None | 1.0 | [0.9697681813033308, 0.03023181869667016] | [0.9697681813033308, 0.03023181869667016] | False | {'fitter': 'linear_inversion', 'fitter_time': ... | 1.0 | True |
| f849e946 | state_fidelity | StateTomography | [Q0] | 0.969727 | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
| 4e995942 | positive | StateTomography | [Q0] | True | None | aer_simulator_from(fake_perth) | None | None | None | None | None | None | None | None |
References¶
See also¶
API documentation:
StateTomography